Overview
As a university breeding program, it is our mandate to conduct high quality peer-reviewed research. The emphasis of our research is on developing and validating new technologies and breeding approaches within the logistical context of an applied breeding program.
Characterizing the diversity in US rice
Although rice has served as a model organism for decades and many genes have been characterized and cloned, much of this work has been conducted outside of elite US germplasm. We leaverage the wealth of genomics data available in the rice community to pinpoint markers for testing in our breeding germplasm. From there we identify key haplotypes and develop SNP assays that can be used for marker assisted selection in our program.

Converting rice knowledge into markers that accelerate the production of new varieties
Our approach has proved fruitful and has led to the development of markers for a multitude of quality, agronomic, and disease traits. Our work includes characterizing the sd1 gene for semidawf stature and exploring BADH2 haplotype variation underlying aroma in US rice.
Speciality rice for Latin markets

PhD student Raul Guerra is working to improve rice for the Latin American export market.
Tastes change over time and it’s our duty to make sure we are developing rice cultivars that not only produce for farmers, but also meet the demands of the consumer. Export to Central America is a key market for US rice farmers; however, exports have declined due changing perceptions of grain quality in these regions. We are working on developing quantitative methods that can be used to understand the grain qualities desired in Latin markets. Once we understand the heritable fraction of these quality traits, we can fastlane the development of varieties that meet quality standards.
Realizing the potential of genome-wide markers
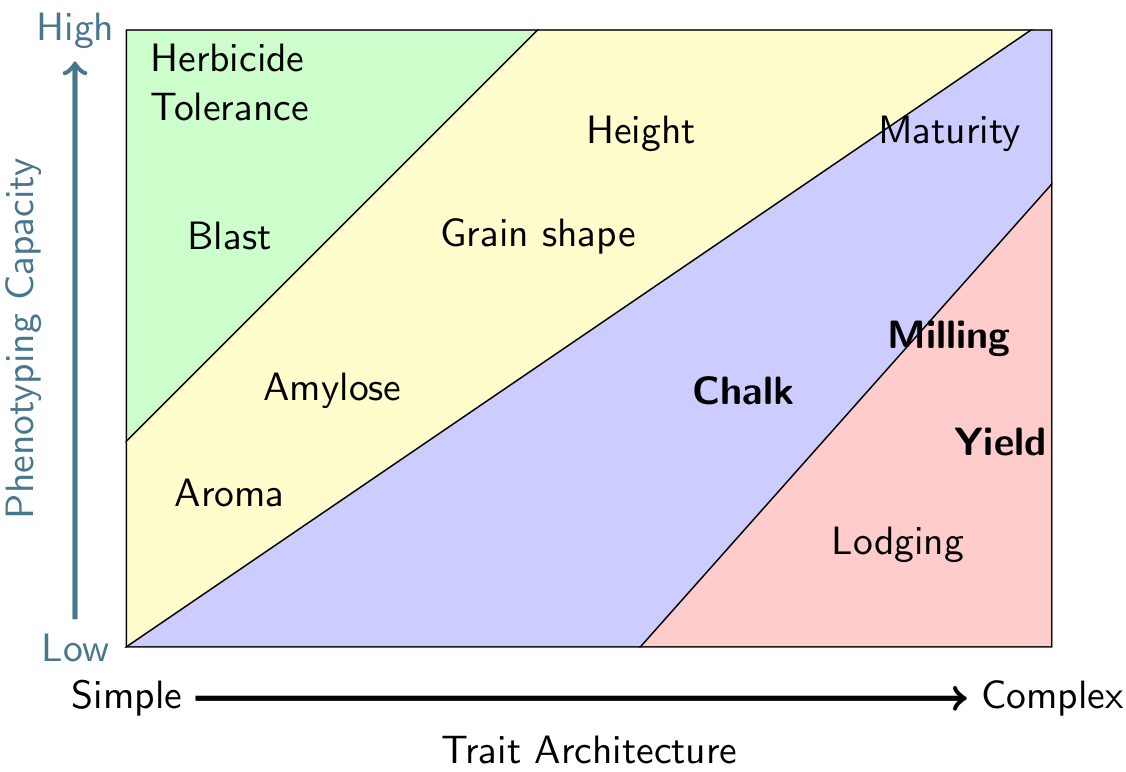
Breeding methods are dictated by phenotyping capacity and genetic complexity. Traits in the bottom right of this chart are prime targets for genomic prediction.
Maker assisted selection can lead to big gains for traits with a simple genetic architecture like plant height, but often fails to deliver for polygenic traits like yield or chalkiness. In the early 2000s, statistical methods for utilizing whole-genome markers to predict complex traits–like yield–were developed and their use has since become mainstream in industry breeding programs for high budget crops. We are investigating the use of genomic prediction within our program in collaboration with the Robbins Lab at Cornell University. read more
Selected research publications
Check out our Google Scholar
Addison, C.K., Angira, B., Cerioli, T., Groth, D.E., Richards, J.K., Linscombe, S.D., and Famoso, A.N. Identification and mapping of a novel resistance gene to the rice pathogen, Cercospora janseana. Theor Appl Genet. 2021.10.1007/s00122-021-03821-2
Cerioli, T., Gentimis, T., Linscombe, S.D, Famoso, A.N. Effect of rice planting date and optimal planting window for Southwest Louisiana. Agronomy Journal. 2021.10.1002/agj2.20593
Addison, C.K., Angira, B., Kongchum, Harrell, D.L., Baisakh, N., Linscombe, S.D., and Famoso, A.N.. Characterization of Haplotype Diversity in the BADH2 Aroma Gene and Development of a KASP SNP Assay for Predicting Aroma in U.S. Rice. 2020. Rice 10.1186/s12284-020-00410-7
Angira, B., Addison, C.K., Cerioli, T., Rebong, D.B., Wang, D.R., Pumplin, N., Ham, J.H., Oard, J.H., Linscombe, S.D. and Famoso, A.N. 2019. Haplotype Characterization of the sd1 Semidwarf Gene in United States Rice. The Plant Genome. 10.3835/plantgenome2019.02.0010